It’s important for non-Bayesians (or non-exclusively-Bayesians) to remember that being a 95% confidence interval procedure is a fairly weak property. It’s not that confidence intervals are necessarily bad, but if they aren’t, it’s because of other requirements.
As an extreme case, consider the all-purpose data-free exact confidence interval procedure for any real quantity: roll a d20 and set the confidence interval to be the empty set if you roll 20, and otherwise to be . That’s a valid confidence interval procedure, it’s just a stupid one.
A much more interesting example comes from a recent paper by Kyle Burris and Peter Hoff, in the context of small-area sampling. First, suppose you have a random variable with known. They say “it is easily verified that” for any function , defines a confidence interval. If you stare at this for a while, you seee that you’re doing a level- test at each value of , but that tells you how to split between upper and lower tails. With you get the standard equal-side test; with you get the lower side only; with you get the upper side only.
What’s unusual is that you get to choose separately for each . That’s fine from the formal definition of confidence intervals: the coverage property only needs to hold for the true , so we need only consider one at a time. But it’s a bit weird.
In the small-area sampling context you have lots of s, s, s, and so on, one for each small area, and you get sets of intervals
From a sampling/frequency point of view, the true are fixed, and depends only on sampling variability. So it’s perfectly ok to allow to depend on data from areas other than area .
In particular, you might have a prior for based on the data from all the other areas, say , and you might want to have shorter confidence intervals when is near . You can do this by biasing the two-sided test to reject on the lower side for and on the upper side for . Specifically, Burris & Hoff define and then and show this all works, and something similar works in the more-plausible setting where you don’t know the s.
In R
make.s<-function(mu, sigma,tau2,alpha){
g<-make.g(alpha)
ginv<-make.ginv(alpha)
function(theta) ginv(2*sigma*(theta-mu)/tau2)
}
make.g<-function(alpha){
function(omega) qnorm(alpha*omega)-qnorm(alpha*(1-omega))
}
make.ginv<-function(alpha){
g<-function(omega) qnorm(alpha*omega)-qnorm(alpha*(1-omega))
function(x){
f<-function(omega) g(omega)-x
if (x<0) {
uniroot(f,c(pnorm(x/alpha),0.5))$root
} else if (x==0) {
0.5
} else
uniroot(f,c(0.5,pnorm(x/alpha)))$root
}
}
ci<-function(y, sigma, s,alpha){
f_low<-function(theta) theta-(y+sigma*qnorm(alpha*(1-s(theta))))
f_up<-function(theta) (y+sigma*qnorm(1-alpha*s(theta)))-theta
low0<-y+qnorm(alpha/2)*sigma
low1<-low0
low<-if (f_low(low1)>0){
while(f_low(low1)>0) low1<- y+(low1-y)*2
uniroot(f_low,c(low1,low0))$root
} else{
uniroot(f_low,c(low0,y))$root
}
hi0<-y+qnorm(1-alpha/2)*sigma
hi1<-hi0
high<-if (f_up(hi1)>0){
while(f_up(hi1)>0) hi1<- y+(hi1-y)*2
uniroot(f_up,c(hi0,hi1))$root
} else{
uniroot(f_up,c(y,hi0))$root
}
c(low,high)
}There’s a sense in which this has got to be cheating. In some handwavy sense, you’re sort of getting away with a confidence interval near at the expense of overcoverage elsewhere. But not really. The intervals really have coverage: if you generated data for any true and used this procedure, of the intervals would cover that true , whatever it was. It’s all good.
But there’s nothing in the maths that requires to be a prior mean. It could also be that is the value you want to convince people that has. Again, you’ll get shorter confidence intervals when they’re centered near your preferred value and longer ones when they aren’t; again, they’ll have coverage
Here’s an example with the true , with , , , and with to make it easier to see. So, we’d like to convince people that , but we aren’t allowed to mess with the 90% confidence level
set.seed(2019-6-11)
s<-make.s(1,1,1,.1)
rr<-replicate(200,{a<-rnorm(1);c(a,ci(a,1, s, 0.1))})
cover<-(rr[2,]<0) & (rr[3,]>0)
plot(1:200, rr[1,],ylim=range(rr),pch=19,col=ifelse(cover,"grey","red"),
ylab=expression(theta),xlab="200 90% confidence intervals")
segments(1:200,y0=rr[2,],y1=rr[3,],col=ifelse(cover,"grey","red"))
abline(h=0,lty=2)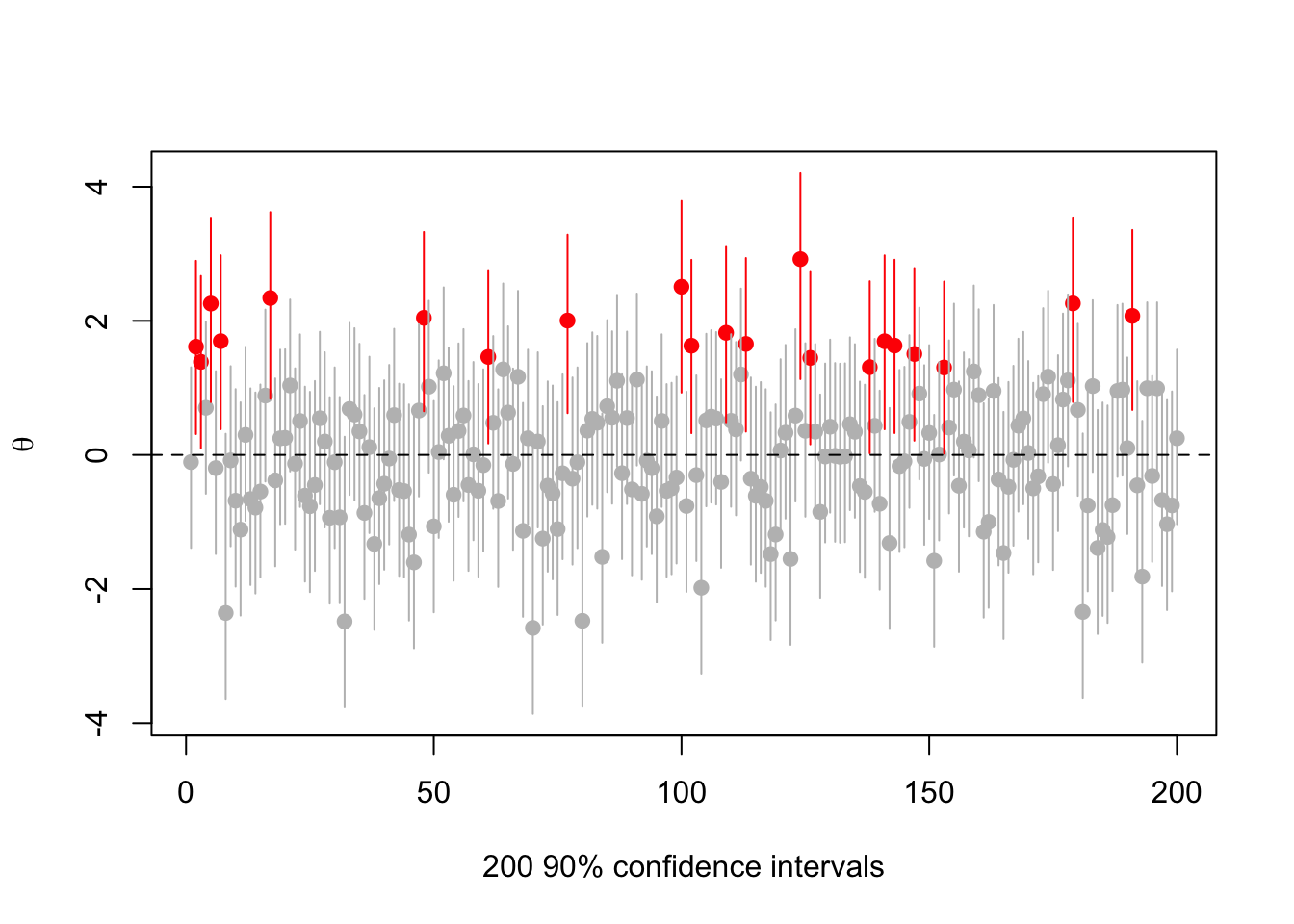
There are 21 intervals that miss zero, consistent with expectations. But they’re all centered at positive .
What happens if we vary the true ? Try (now the red intervals are those that don’t cover 1)
set.seed(2019-6-11)
s<-make.s(1,1,1,.1)
rr<-replicate(200,{a<-rnorm(1,1);c(a,ci(a,1, s, 0.1))})
cover<-(rr[2,]<1) & (rr[3,]>1)
plot(1:200, rr[1,],ylim=range(rr),pch=19,col=ifelse(cover,"grey","red"),
ylab=expression(theta),xlab="200 90% confidence intervals")
segments(1:200,y0=rr[2,],y1=rr[3,],col=ifelse(cover,"grey","red"))
abline(h=1,lty=2)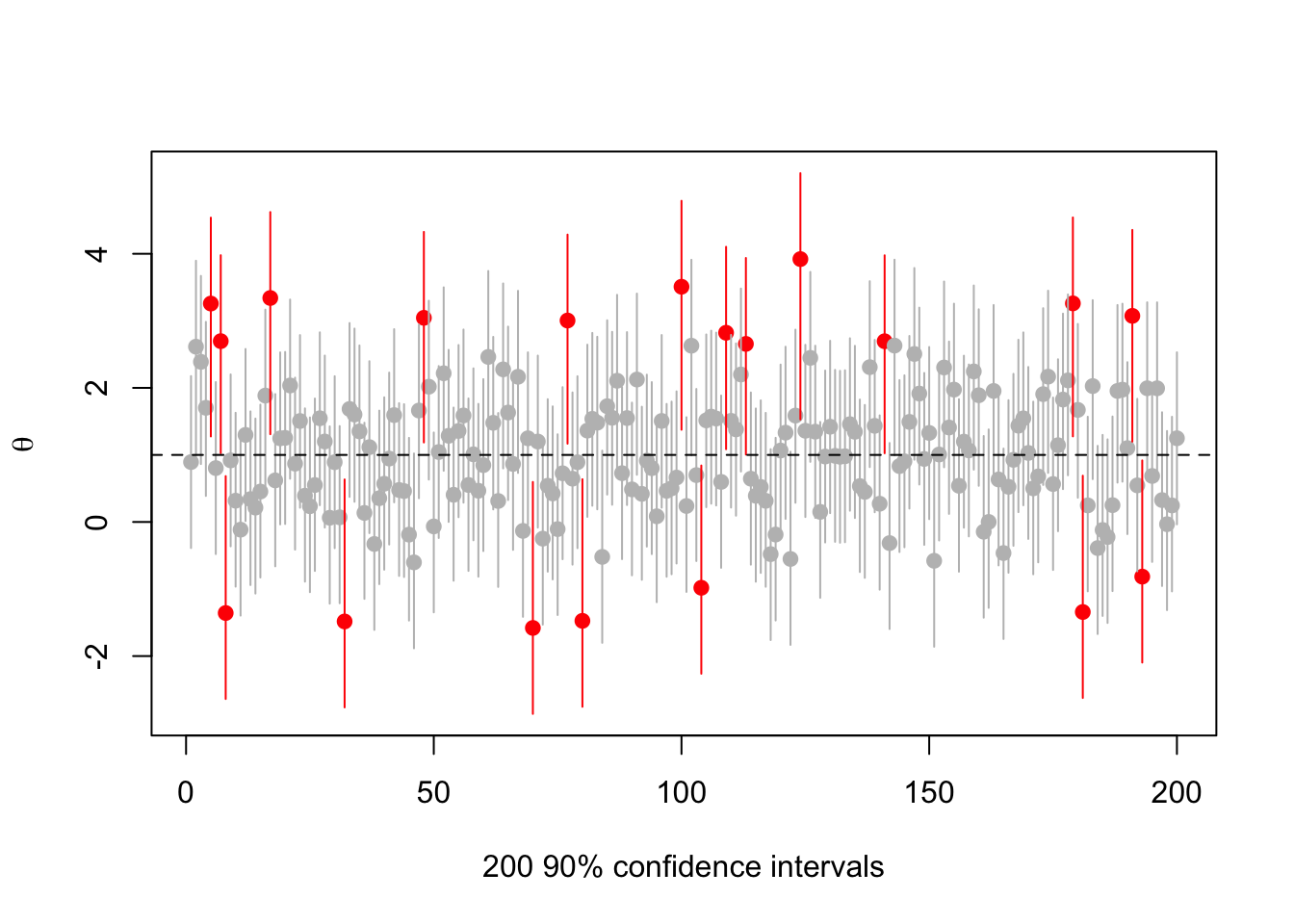
Or
set.seed(2019-6-11)
s<-make.s(1,1,1,.1)
rr<-replicate(200,{a<-rnorm(1,2);c(a,ci(a,1, s, 0.1))})
cover<-(rr[2,]<2) & (rr[3,]>2)
plot(1:200, rr[1,],ylim=range(rr),pch=19,col=ifelse(cover,"grey","red"),
ylab=expression(theta),xlab="200 90% confidence intervals")
segments(1:200,y0=rr[2,],y1=rr[3,],col=ifelse(cover,"grey","red"))
abline(h=2,lty=2)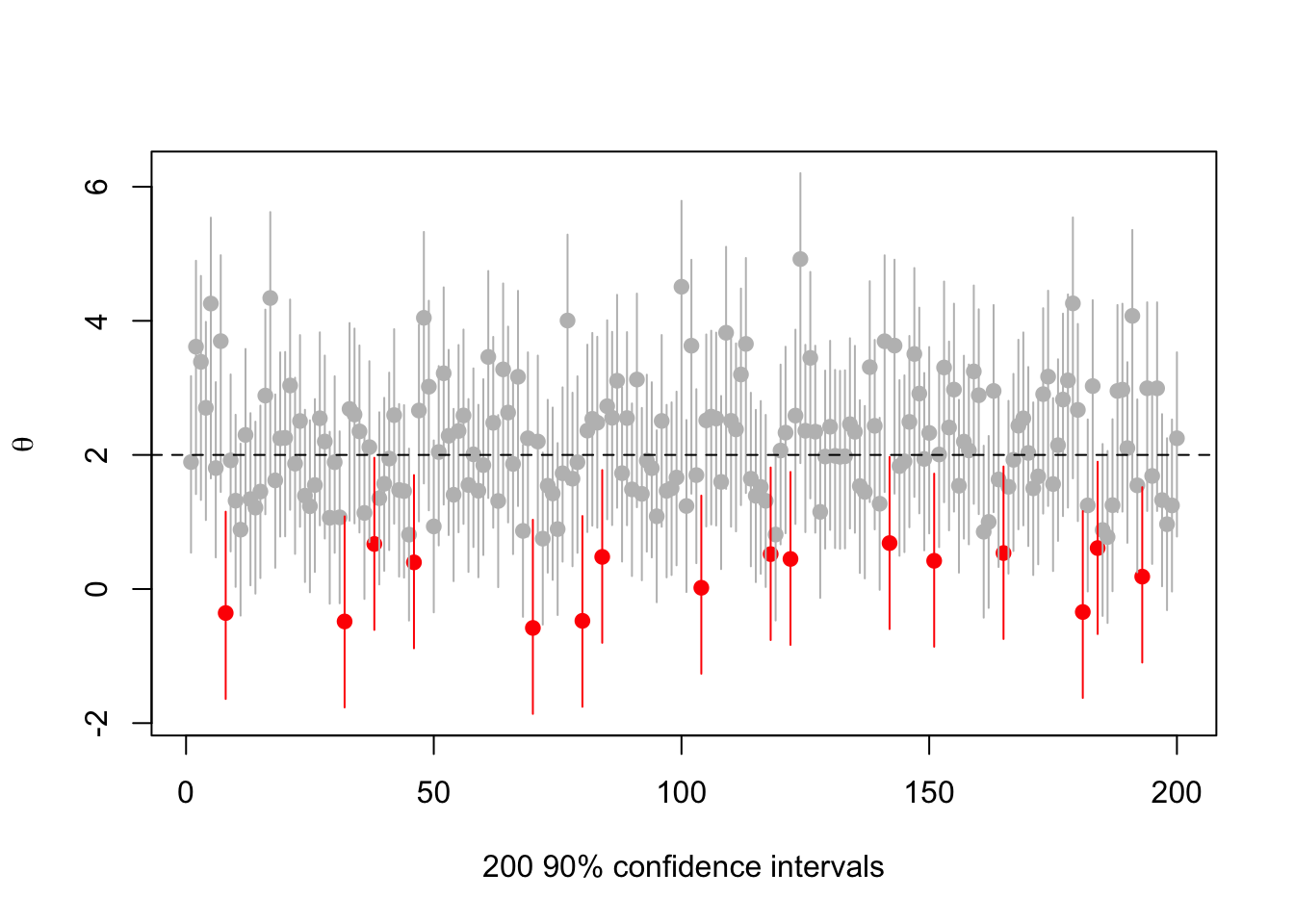
Or
set.seed(2019-6-11)
s<-make.s(1,1,1,.1)
rr<-replicate(200,{a<-rnorm(1,-1);c(a,ci(a,1, s, 0.1))})
cover<-(rr[2,]< -1) & (rr[3,]> -1)
plot(1:200, rr[1,],ylim=range(rr),pch=19,col=ifelse(cover,"grey","red"),
ylab=expression(theta),xlab="200 90% confidence intervals")
segments(1:200,y0=rr[2,],y1=rr[3,],col=ifelse(cover,"grey","red"))
abline(h=-1,lty=2)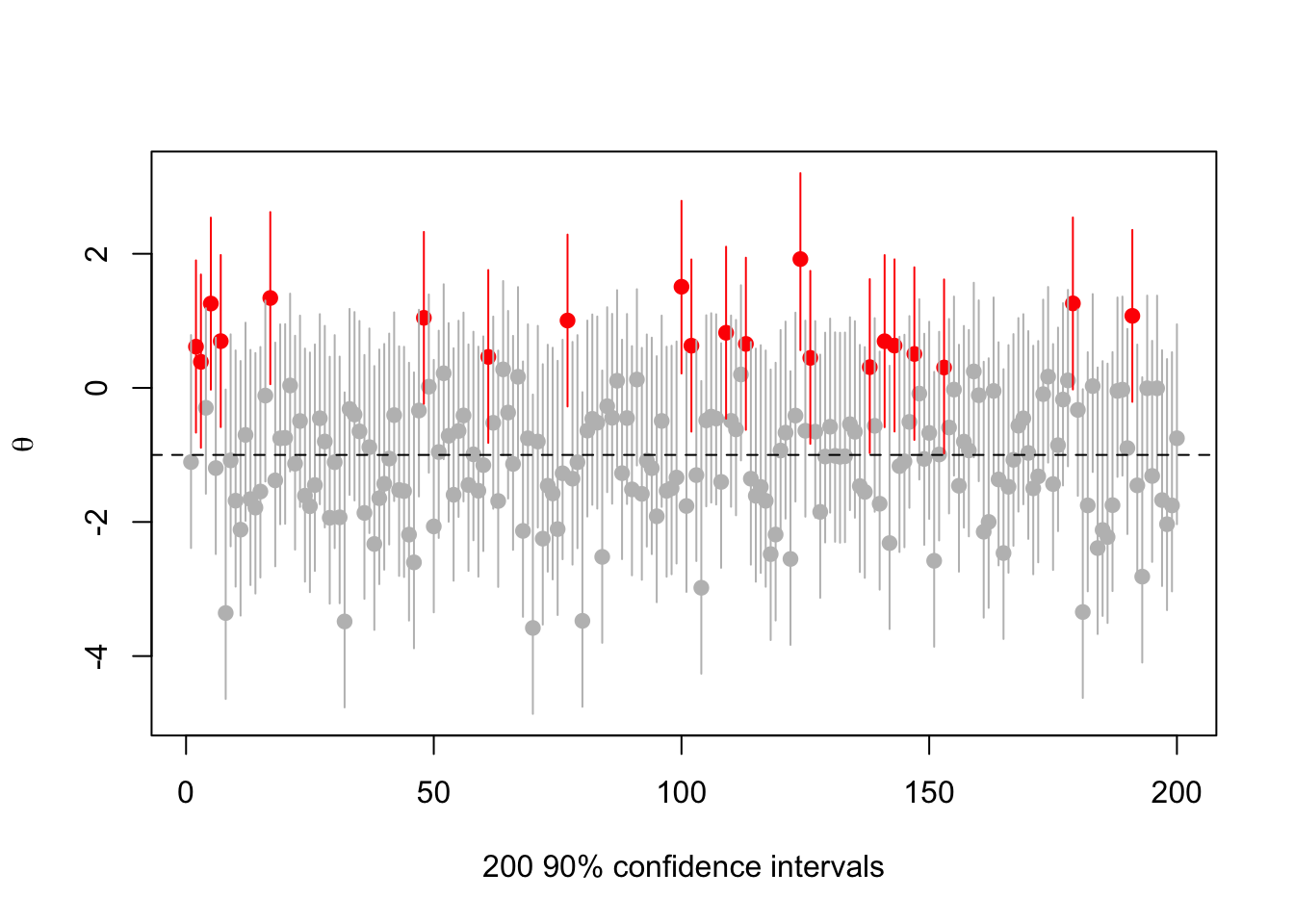
There’s nothing actually wrong here, but it’s messing with people’s natural misunderstandings of confidence intervals in order to push the impression that .
I think I like it. Or maybe I don’t. One or the other.